SCReadCounts Examples
Command-line:
Example: BAM files and single SNV file in TSV format.
sh singlecell.sh
The above script runs SCReadCounts using the inputs from SCReadCounts/data (snv file = singlecell_222_5_chr17.txt, bam = singlecell_chr17.bam, output prefix = singlecell-output.tsv). Result files corresponding to this analysis are available here
Example output files:
Graphical User Interface
Example 1: BAM files and single SNV file in txt format with default minR value (5).
-
Select the SNV file by clicking on the
Browsebutton, navigating toSCReadCounts/data, selectingsinglecell_222_5_chr17.txt, and clickingOK. -
Select the BAM files by clicking on the
Browsebutton, navigating toSCReadCounts/data, selectingsinglecell_chr17.bam(multiple BAM files can be selected using shift-click or control-click as needed), and clickingOK. -
Specify the filtering option by clicking on the
🠋button.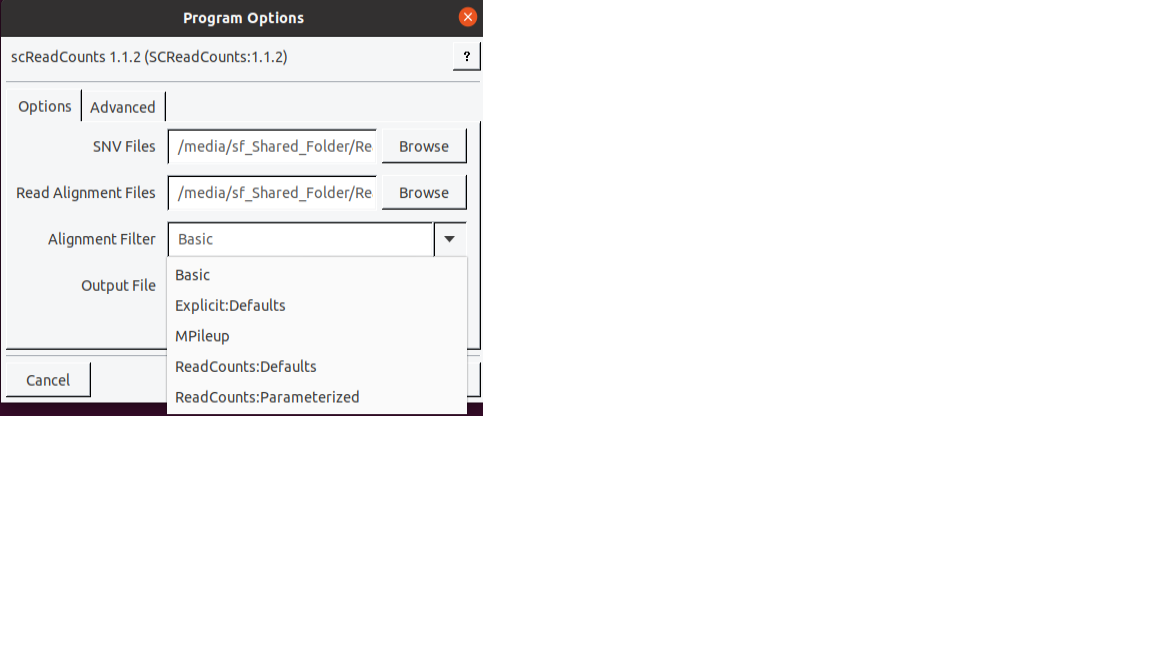
-
Specify the output directory by clicking on the
Browsebutton, navigating toSCReadCounts/data, clickingCreate Folder, entering “new_output”, entering the name assinglecell_GUI.tsvand clickingSave.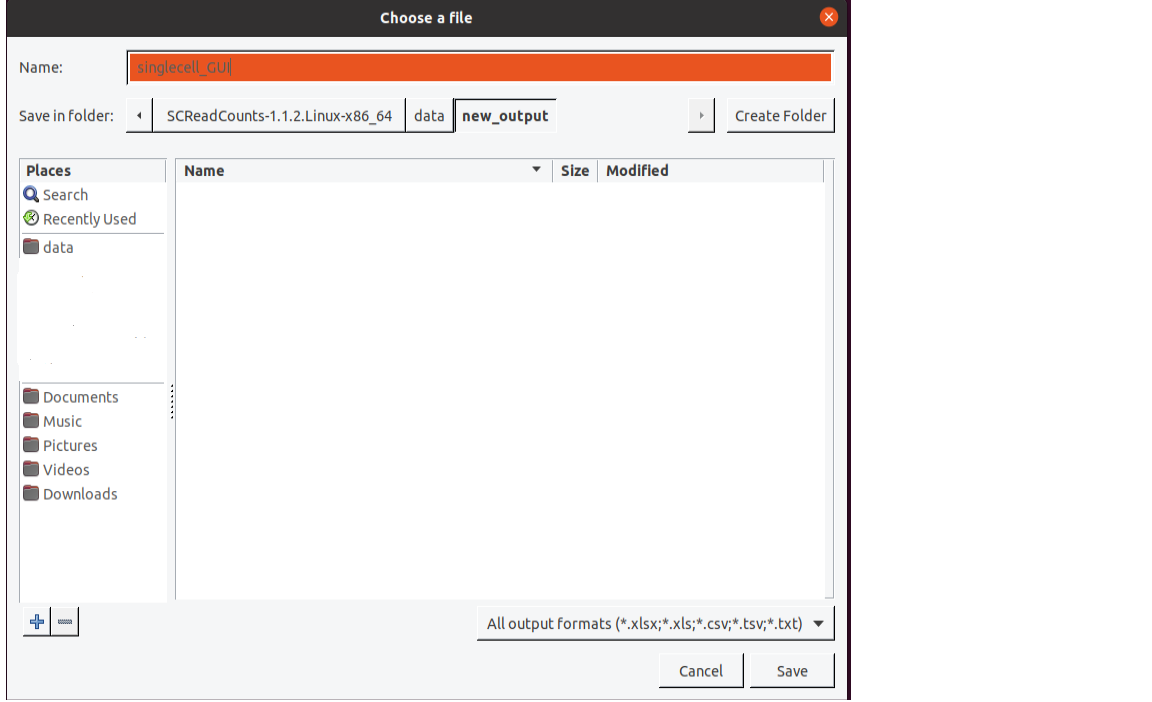
SCReadCounts will create the following files:
- Counts(singlecell_GUI.tsv)
- Count matrix (Ref;Var)(singlecell_GUI.cnt.matrix.tsv)
- VAF matrix with minR=5 (singlecell_GUI.vaf-m5.matrix.tsv)
- Click
OKto execute the program.
Example 2: Re-running SCReadCounts with different minR value (10).
-
Select the SNV file by clicking on the
Browsebutton, navigating toSCReadCounts/data, selectingsinglecell_222_5_chr17.txt, and clickingOK. -
Select the BAM files by clicking on the
Browsebutton, navigating toSCReadCounts/data, selectingsinglecell_chr17.bam(multiple BAM files can be selected using shift-click or control-click as needed), and clickingOK. -
Specify the filtering option by clicking on the
🠋button. -
Specify the output directory by clicking on the
Browsebutton, navigating toSCReadCounts/data, clickingCreate Folder, entering “new_output”, entering the name assinglecell_GUI.tsvand clickingSave. You will be prompted with a messageA file named "singlecell_GUI.tsv" already exists. Do you want to replace it?. Click on theReplacebutton.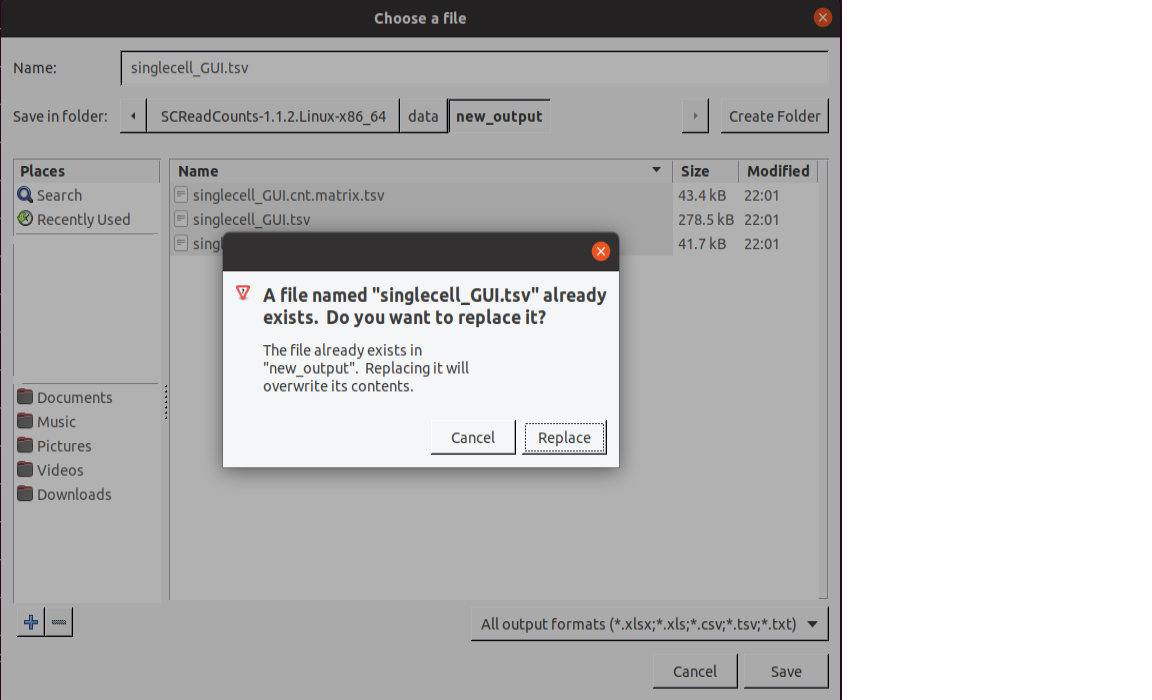
-
Click
OKto execute the program.
Since the output files: singlecell_GUI.tsv and singlecell_GUI.cnt.matrix.tsv already exists (it doesn’t change with respect to the “advanced” option of minR), only one new file will be created i.e. VAF with minR of 10 (singlecell_GUI.vaf-m10.matrix.tsv)