RNA2DNAlign Examples
Command-line:
Example 1: BAM files and single SNV file in TSV format.
cd RNA2DNAlign/data
../bin/RNA2DNAlign -r "example-*.bam" -s "example-SNV.tsv" -o example1
Example 2: BAM and VCF files for each dataset, exonic SNV filtering, and DARNED and COSMIC annotations using the supplied annotation files.
cd RNA2DNAlign/data
../bin/RNA2DNAlign -r "example-GDNA.bam example-NRNA.bam example-SDNA.bam example-TRNA.bam" -s "example-*.vcf" -o example2 -e UCSC_Human_hg19_RefSeq_CDS_exon_coordinates.txt -d DARNED_hg19.txt -c CosmicMutantExport_hg19.tsv.gz
Result files corresponding to this analysis are available in the RNA2DNAlign/data directory in the example-output directory.
Example output files:
- Events_LOH.tsv
- Events_RNAed.tsv
- Events_SOM.tsv
- Events_T-RNAed.tsv
- Events_T-VSE.tsv
- Events_T-VSL.tsv
- Events_VSE.tsv
- Events_VSL.tsv
- readCounts.tsv
- summary_result.txt
Example 3: BAM and VCF files for each dataset, exonic SNV filtering, and minimum reads per loci in each dataset of 3.
cd RNA2DNAlign/data
../bin/RNA2DNAlign -r "example-*.bam" -s "example-*.vcf" -o example3 -e UCSC_Human_hg19_RefSeq_CDS_exon_coordinates.txt -m 3
Graphical User Interface
Example 1: BAM files and single SNV file in TSV format.
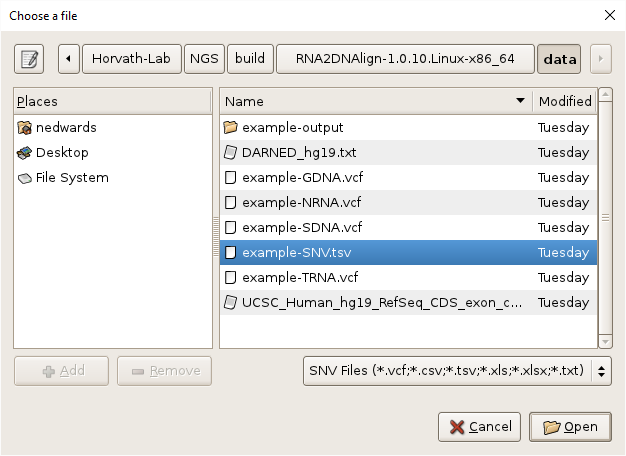
- Select the SNV file by clicking on the
Browsebutton, navigating toRNA2DNAlign/data, selectingexample-SNV.tsv, and clickingOK.
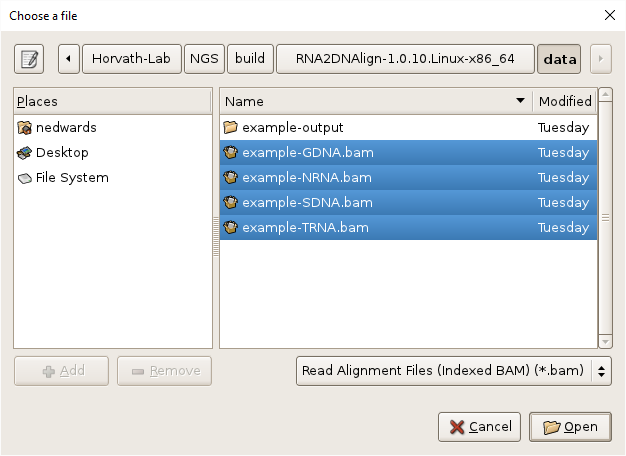
- Select the BAM files by clicking on the
Browsebutton, navigating toRNA2DNAlign/data, selecting all the BAM files, using shift-click or control-click as needed, and clickingOK.
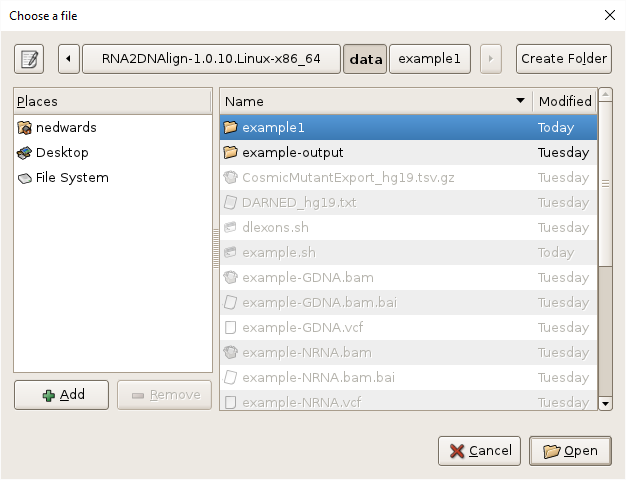
- Specify the output directory by clicking on the
Browsebutton, navigating toRNA2DNAlign/data, clickingCreate Folder, entering “example1”, and clickingOpen.
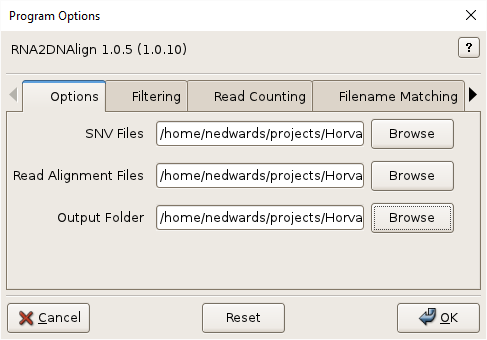
- Click
OKto execute the program.
Example 2: BAM and VCF files for each dataset, exonic SNV filtering, and DARNED and COSMIC annotations using the supplied annotation files.
- Select the VCF files by clicking on the
Browsebutton, navigating toRNA2DNAlign/data, selecting all the VCF files, using shift-click or control-click as needed, and clickingOK.
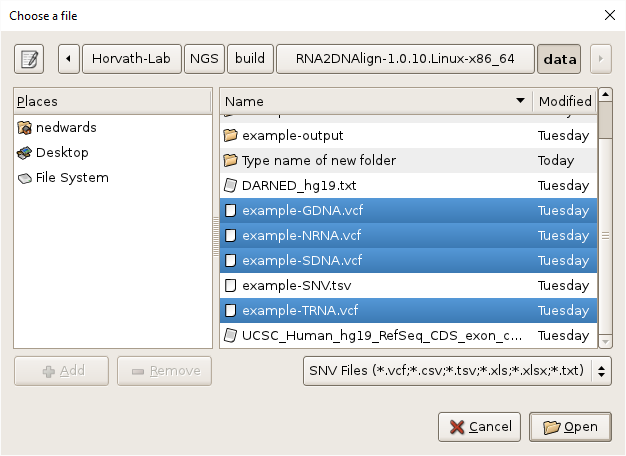
- Select the BAM files by clicking on the
Browsebutton, navigating toRNA2DNAlign/data, selecting all the BAM files, using shift-click or control-click as needed, and clickingOK.

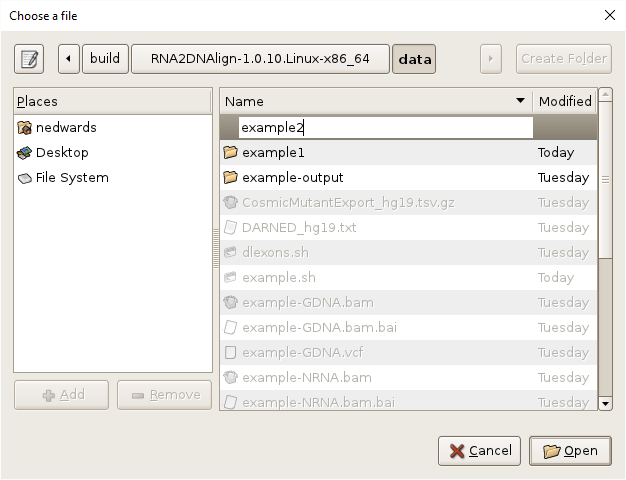
- Specify the output directory by clicking on the
Browsebutton, navigating toRNA2DNAlign/data, clickingCreate Folder, entering “example2” and clickingOpen.
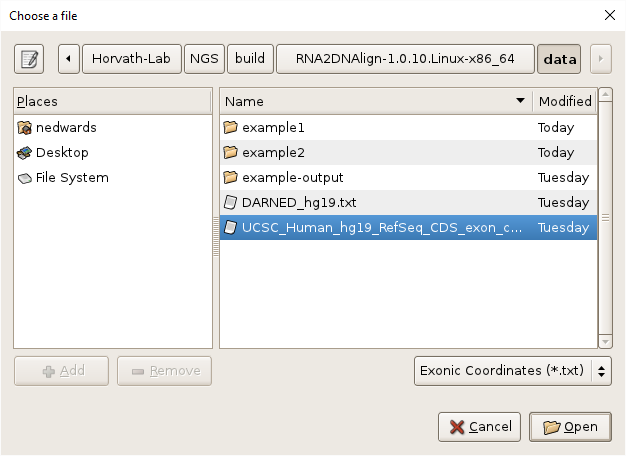
- Specify exonic SNV filtering by selecting the
Filteringtab, clicking on theBrowsebutton, navigating toRNA2DNAlign/data, selecting “UCSC_Human_hg19_RefSeq_CDS_exon_coordinates.txt”, and clickingOK.
-
Specify DARNED and COSMIC annotation of SNP events on the
SNV Annotationtab, selecting the filesDARNED_hg19.txtandCosmicMutantExport_hg19.tsv.gz. -
Click
OKto execute the program.
Result files corresponding to this analysis are available in the RNA2DNAlign/data directory in the example-output directory.
Example output files:
- Events_LOH.tsv
- Events_RNAed.tsv
- Events_SOM.tsv
- Events_T-RNAed.tsv
- Events_T-VSE.tsv
- Events_T-VSL.tsv
- Events_VSE.tsv
- Events_VSL.tsv
- readCounts.tsv
- summary_result.txt
See Also
RNA2DNAlign Home, Input Files, Output Files, Annotation Files